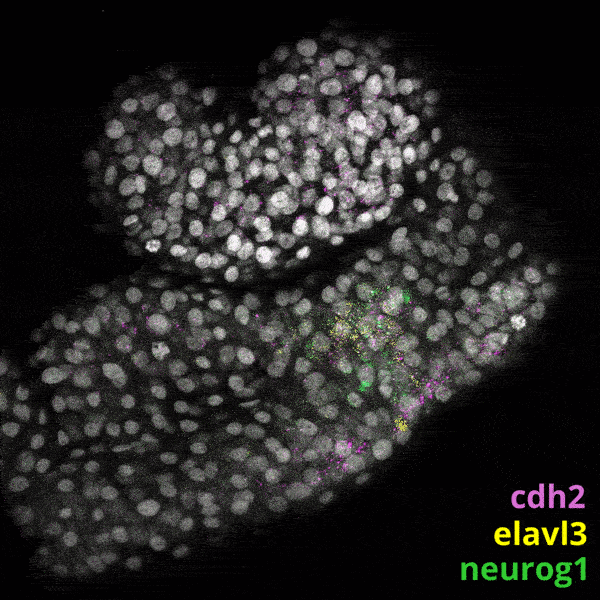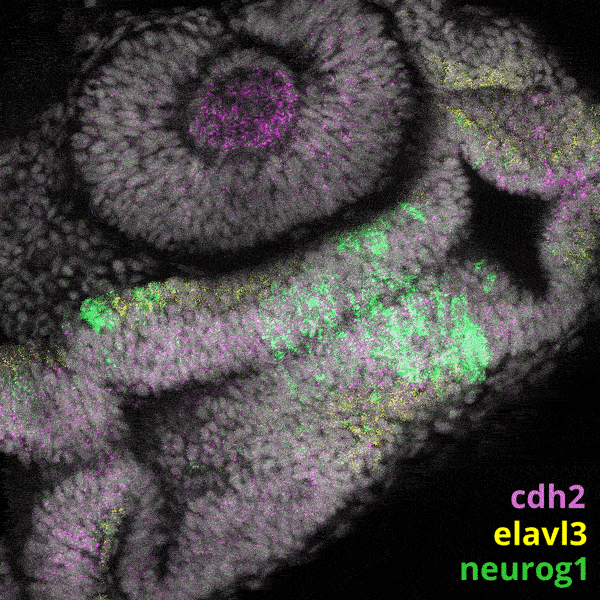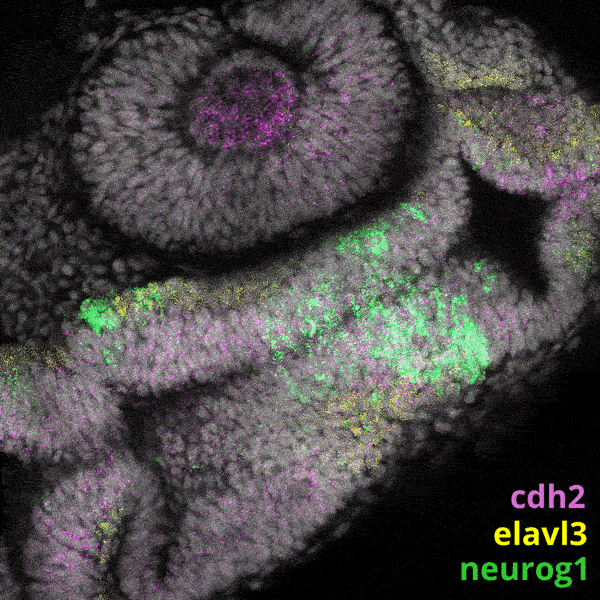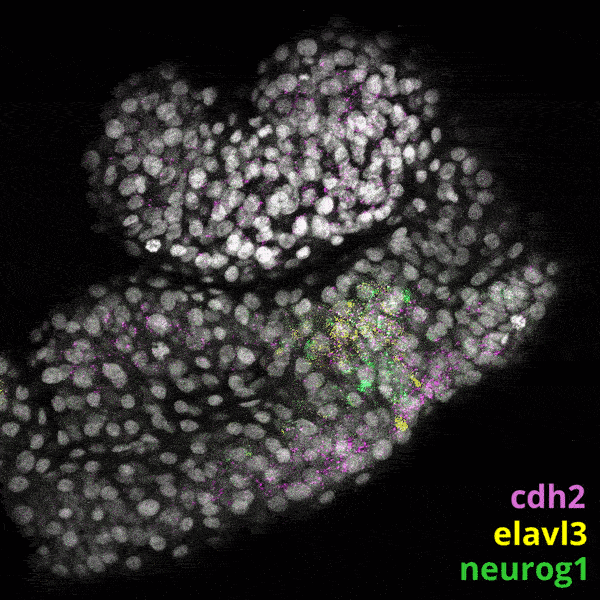
Zebrafish Development
We experiment primarily with zebrafish (Danio rerio), a powerful molecular genetic model system for vertebrate biology. Zebrafish develop rapidly outside the mother’s body and are transparent at early stages, enabling the tracking and manipulation of cells in living embryos. Zebrafish share many of the same organs and genes as humans, making this an ideal system for rapid interrogations of developmental defects and genetic disease. Routine access to hundreds of embryos from any developmental stage of interest allows us to greatly scale in situ hybridizations, molecular perturbations, and genomics experiments.
Single-Cell ‘omics
We utilize microfluidic technologies to process cells from a range of life stages for high-throughput single-cell genomics. inDrops is an open-source and customizable microfluidic droplet platform capable of encapsulating tens of thousands of cells per hour for single-cell RNA sequencing.
Quantitative Developmental Biology
We have demonstrated a “shotgun” approach for reconstructing quantitative maps of embryonic development from tens of thousands of single-cell gene expression snapshots. These snapshots are collected at random, but can be “stitched” together into state manifolds that depict gene expression trajectories for all organs and tissues of the developing embryo.
We have built and annotated a state manifold encompassing the first 24 hours of zebrafish development. We are mining this resource to identify novel candidate genes that regulate the specification of key tissues and organs. Importantly, this manifold contains several features that are inconsistent with a simple tree-like topology, for example, converging trajectories (or “loops”), in which distinct paths ultimately lead to same final cell state. We are studying such trajectories to gain insights into natural examples of cellular transdifferentiation.
High-Throughput Lineage Tracing
Lineage hierarchies reflect the mitotic history of a tissue and can be used to identify cell fate restriction events in a differentiation cascade. Traditional imaging-based methods, however, can only pair lineage information with a limited number of state measurements per cell. We have developed a sequencing-based method called Tracer-Seq, which combines lineage tracing with simultaneous high-dimensional transcriptome measurements in single cells. We use Tracer-Seq to study how lineage hierarchies unfold on a state manifold, and to independently verify the accuracy of predicted gene expression trajectories.
Perturbation Mapping
Single-cell state manifolds can reveal detailed gene expression information for all tissues of an animal or embryo simultaneously. We leverage state manifolds to map developmental patterning phenotypes that manifest distinct molecular defects in different tissues of the body, and/or which alter the proportions of otherwise normal tissues.
Our goals include:
To identify experimental strategies for controlling how cells “flow” through the state manifold.
To identify how and when pathological cell states emerge.
To characterize regulative feedback mechanisms that maintain normal development.
To combine comparative analyses with machine learning to characterize human developmental diseases and birth defects.